

vseed.nottingham.ac.uk
This site contains web-based tools for analysis of gene expression occurring during seed maturation, dormancy and germination.
Related Resources:
Online Network Query
Tools:

SeedNet - Genome-wide transcription
network in seeds (original reference: Bassel et
al. PNAS (USA) 2011):
A queryable interface for SeedNet, a genome-wide network
model describing transcriptional interactions in dormant
and germinating Arabidopsis seeds.
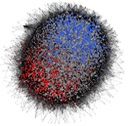 Seed Co-Prediction Network - SCoPNet
for seed dormancy and germination (original
reference: Bassel et al. The Plant Cell,
2011):
Seed Co-Prediction Network - SCoPNet
for seed dormancy and germination (original
reference: Bassel et al. The Plant Cell,
2011):
A queryable interface for SCoPNet, a genome-wide network
model describing transcriptional interactions in
germinating and dormant Arabidopsis seeds, generated
through machine learning.
People:


 Micropylar Endosperm-(END)net
(original reference: Dekkers et al. Plant
Physiology 2013):
Micropylar Endosperm-(END)net
(original reference: Dekkers et al. Plant
Physiology 2013):
A queryable interface for ENDnet, a genome-wide network
model describing transcriptional interactions in the
micropylar endosperm of germinating Arabidopsis seeds.
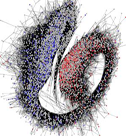 Radicle-(RAD)net (original reference:
Dekkers
et al. Plant Physiology 2013):
Radicle-(RAD)net (original reference:
Dekkers
et al. Plant Physiology 2013):
A queryable interface for RADnet, a genome-wide network
model describing transcriptional interactions in the lower
axis of germinating Arabidopsis seeds.

Gene expression
visualisations
(eFP browsers):
 High
resolution time-course course of germination in
Arabidopsis (original
reference: Dekkers et al. Plant Physiology
2013)
High
resolution time-course course of germination in
Arabidopsis (original
reference: Dekkers et al. Plant Physiology
2013)
An eFP Browser of seed germination, analysing expression
in separated radicle, cotyledon, lateral and micropylar
endosperm.
Other resources:
 TAGGIT
analysis of seed transcriptome datasets: (original
reference: Carrera et al Plant Physiology 2007):
TAGGIT uses a seed germination/dormancy ontology to
annotate 'omics datasets, thus allowing more biologically
informed analysis. A guide and most up-to-date version
(although not updated since 2009, you can also use your
own gene lists) of the TAGGIT workflow.
TAGGIT
analysis of seed transcriptome datasets: (original
reference: Carrera et al Plant Physiology 2007):
TAGGIT uses a seed germination/dormancy ontology to
annotate 'omics datasets, thus allowing more biologically
informed analysis. A guide and most up-to-date version
(although not updated since 2009, you can also use your
own gene lists) of the TAGGIT workflow.
vSEED Web
Portal:
Web portal for the ERANET-PG funded vSEED project.
The
vSEED Web Resource is associated with:
The ERANET-PG vSEED
project and Infobiotics (University of
Nottingham).
This project is a collaboration between Professor
Michael Holdsworth and Professor Nat
Krasnogor, School
of Computer Science; University of
Nottingham.
Funding for this project was
originally from ERANET-PG
(BB/G02488X/1), BBSRC.
Site managed by Professor
Michael Holdsworth with a great deal of help from
MyCIB.
Last update 01 March 2013.







